Li Bowen of the University of Toronto and Daniel Anderson of the Massachusetts Institute of Technology published a research paper entitled “Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry” in the Nature sub-journal Nature Materials.
By combining machine learning and combinatorial chemistry to establish a method to accelerate the discovery and evaluation of ionizable lipids, ionizable lipids 119-23 were identified as superior to established baseline lipids in transfecting muscle and immune cells from several tissues, leading to the development of lipid nanoparticles (LNPs) for precise delivery of mRNA.
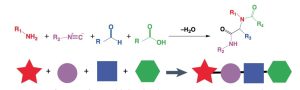
Fig. 1 A 4CR for HTS of Ionizable Lipids1
Lipid nanoparticles (LNPs) have made substantial clinical progress over the past few years, the first siRNA drug from Alnylam in 2018 (for the treatment of peripheral neuropathy caused by hereditary transthyretin amyloidosis), and two COVID-19 mRNA vaccines from Moderna and Pfizer/BioNTech in 2020. Notably, all three LNP formulations contain a unique ionizable lipid. The design space for ionizable lipids is vast, and small changes in their chemistry can greatly affect biological function.
Daniel Anderson and Bowen Li’s team has been exploring high-throughput synthesis (HTS) strategies to accelerate the design and identification of novel ionizable lipids for RNA delivery. Only a few chemical reactions are compatible with the thermodynamic stability of lipids in biocompatible solvents and can be performed without the use of catalysts or cumbersome protection/deprotection steps. Two notable examples include the Michael addition reaction, which is the 1,4-addition of amines to acrylates, and the ring-opening reaction of amines to epoxides.
However, due to their two-dimensional structure, these reactions have inherent limitations in terms of the diversity and structural flexibility of ionizable lipids that can actually be generated. To overcome this limitation, Daniel Anderson and Bowen Li’s team published a paper in the journal Nature Biotechnology in March 2023, detailing the design of a three-component reaction (3CR) system, In this system, a nitroricinoleate acrylate (NRA) linker is coupled to a fatty alcohol (lipid tail) and then linked to a head group containing primary, secondary, or tertiary amines. Compared with the traditional two-component reaction, where the amine head group is directly bound to the lipid tail, the 3CR system simplifies the tedious synthesis process, increases lipid structural diversity, and rapidly generates a structurally diverse biodegradable ionized lipid portfolio library. On this basis, RCB-4-8 LNP was screened and identified, showing better biodegradability and safety. It can be reproducibly administered intratracheally and, for the first time, successfully used LNP to achieve efficient CRISPR gene editing in lung epithelial cells, opening up new possibilities for gene therapy for congenital lung diseases.
In this latest study, published in the journal Nature Materials, the research team constructed a novel high-throughput synthesis (HTS) platform based on a four-component reaction (4CR) to make the design and generation of ionizable lipids more efficient. Conceptually, the ionizable lipid structure is divided into four independent elements: amine head group, linker, tail 1 and tail 2, corresponding to the amines, isocyanates, aldehydes and carboxylic acids in the 4CR reactants, respectively. This 4CR method is an improvement over the previous 3CR method, allowing for increased dimensionality and yields, which facilitates the generation of more ionizable lipid candidates.
This study developed an approach that combines machine learning with cutting-edge combinatorial chemistry tools to accelerate the discovery of potent ionizable lipids for mRNA delivery. Starting with a simple four-component reaction (4CR) platform, the research team created a chemically diverse library of 584 ionizable lipids. The mRNA transfection efficiency of lipid nanoparticles (LNPs) containing these ionizable lipids was screened and used as a basic dataset to train various machine learning models.
The team then selected the best-performing machine learning model to explore a vast virtual lipid library of 40,000 lipids to synthesize and experimentally evaluate the top 16 ionizable lipids. The research team found ionizable lipid 119-23, which outperformed established baseline lipids in transfecting muscle and immune cells from several tissues.
Overall, this study facilitates the development of lipid nanoparticles (LNPs) for precise delivery of mRNA by combining machine learning and combinatorial chemistry to establish a method to accelerate the discovery and evaluation of ionizable lipids.
See more mRNA delivery services:
- Lipid-based Vectors for mRNA Delivery
- Polymer-based Vectors for mRNA Delivery
- Hybrid Vectors for mRNA Delivery
Reference
- Li, Bowen, et al. “Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry.” Nature Materials(2024): 1-7.
