The concentration of proteins containing intrinsically disordered regions (IDRs) must be tightly controlled to maintain cellular homeostasis. However, our understanding of the collective control mechanisms for these proteins—which tend to cluster in membraneless condensates—is far less comprehensive than that of pathways mediated by membrane-bound organelles.
Recently, researchers from the Francis Crick Institute and King’s College London published a research paper in the top-tier international academic journal Nature, titled: Collective homeostasis of condensation-prone proteins via their mRNAs.
The study reveals a complex yet exquisite homeostatic regulatory mechanism within the cell nucleus called “interstasis.” This mechanism coordinates the expression of proteins prone to forming biomolecular condensates via phase separation. When the concentration of these proteins becomes too high, it triggers their own mRNAs to be “captured” and sequestered within nuclear speckles, preventing their further translation into more protein and ultimately reducing the protein’s concentration. Conversely, when the protein concentration falls, the negative feedback loop is switched off. The study further reveals the regulatory switch for this circuit. This negative feedback loop adds a new dimension to the function of nuclear speckles, highlighting the interplay between RNA sequence characteristics and protein domain architecture in cellular regulation.
Adaptive screening across multiple eukaryotic species has shown that proteins containing intrinsically disordered regions (IDRs) are more likely to be toxic when their dosage is increased. In yeast, the toxicity induced by the dosage increase of a single IDR-containing protein has been linked to the tendency of these proteins to undergo phase separation and form biomolecular condensates.
Proteins prone to phase separation exhibit the greatest mismatch between their protein and RNA abundances, suggesting a strong influence of post-transcriptional mechanisms on protein levels. While many proteins are capable of interacting with the RNA or DNA of their own genes, there have been no known sensors or effectors capable of regulating the collective expression of condensation-prone proteins.
The co-condensation of proteins depends, to some extent, on the molecular grammar of short motifs within their IDRs and on amino acids with similar biophysical properties. Many IDRs contain repetitive amino acid arrangements and are therefore classified as low-complexity domains (LCDs). One class of LCDs is the R-MCD (arginine-mixed charge domain), where arginine is interspersed with other positively and negatively charged amino acids. R-MCDs promote the localization of proteins to nuclear speckles.
Studies have shown that significant overexpression of artificial R-MCDs enhances the cohesion of nuclear speckles, leading to the retention of all polyadenylated mRNAs within them. This observation underscores the importance of coordinating the homeostatic regulation of this class of proteins.
The study discovered a complex yet exquisite mechanism of cellular homeostasis—interstasis—a collective negative feedback mechanism of intracellular homeostasis. When the concentration of a specific protein becomes too high within an RNA-protein membraneless condensate (such as nuclear speckles), it triggers its own mRNA to be “captured” and sequestered within that condensate, thereby preventing its translation into more protein and ultimately reducing the protein’s concentration.
Nuclear speckles, a typical biomolecular condensate, become overly cohesive, or “too sticky,” when there is an excess of R-MCD-rich proteins. This change in physical state acts as a “sensor,” detecting the signal of excessive protein concentration. These “too sticky” nuclear speckles then begin to selectively capture the mRNAs that encode condensation-prone proteins.
This selectivity is based on the structure of the genetic code and conserved codon usage preferences. For instance, mRNAs encoding R-MCDs are often themselves rich in purine (AG-rich) repeats, forming multivalent regions. This structural similarity allows them to be recognized and bound by specific RNA-binding proteins (e.g., TRA2 protein) that also accumulate in the nuclear speckles. The captured mRNAs are retained inside the nuclear speckles, unable to exit to the cytoplasm for ribosomal translation, which leads to reduced synthesis and concentration of the corresponding protein. Once the protein concentration drops, the cohesion of the nuclear speckles returns to normal, and the negative feedback loop is switched off.
The research team further revealed the regulatory switch for interstasis: CLK kinase-mediated phosphorylation of the TRA2 protein can prevent its localization to the nuclear speckles. This means that phosphorylation inhibits interstasis, preventing the feedback loop from starting. This provides the cell with a fine-tuning “switch” for this mechanism.
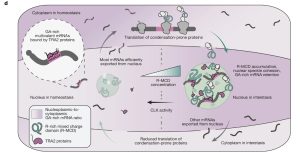
Fig.1 Schematic detailing the interstasis of proteins with charged low-complexity domains.1
These findings solve a key problem: explaining how cells collectively control the expression of a series of highly dose-sensitive, condensation-prone proteins, preventing their overexpression from leading to uncontrolled phase separation and the formation of toxic aggregates, which can cause cellular dysfunction or diseases (such as neurodegenerative disorders). These discoveries also reveal a completely new regulatory dimension—cleverly linking the biochemical properties of the protein (phase separation capacity), the sequence characteristics of the mRNA (codon bias), and the physical state of the subcellular structure (condensate viscosity), forming a rapid response mechanism that transcends traditional transcriptional regulation.
In summary, Interstasis is an extremely clever mechanism of cellular homeostasis: the cell uses the condensate itself as a sensor, and through changes in its physical state, directly recruits and silences the mRNAs of proteins that would “cause trouble” if overabundant, thereby achieving collective self-maintenance of homeostasis.
Creative Biolabs Custom mRNA Modification: Enhanced Therapeutics
Creative Biolabs offers comprehensive Custom mRNA Modification Services to significantly enhance the stability, reduce immunogenicity, and boost the translational efficiency of synthetic mRNA for therapeutic and research applications.
Key Modification Offerings:
- mRNA 5′ Capping: The 5′ cap is crucial for efficient translation and stability. We provide advanced capping options:
- ARCAs Cap (Anti-Reverse Cap Analog): Ensures high yield of correctly oriented, translatable mRNA.
- Fluorescent Cap: Ideal for tracing and localization studies.
-
- Fluorophosphate-containing Cap and 6-Thioguanosine-containing Cap: Used for specific biochemical and structural research applications.
- 2. Nucleotides Modification: Incorporating modified nucleosides is a powerful strategy to minimize host immune response and prolong the mRNA’s functional half-life. Our available modifications include:
-
- Pseudouridine Modification
- 2-Thiouridine Modification
- 5-Methylcytidine Modification
- N6-Methyladenosine Modification
-
By strategically implementing these modifications, Creative Biolabs helps researchers unlock the full potential of mRNA-based therapeutics and vaccines.
Reference
- Faraway, Rupert, et al. “Collective homeostasis of condensation-prone proteins via their mRNAs.” Nature(2025): 1-11. CC BY 4.0. https://doi.org/10.1038/s41586-025-09568-w
