Influenza A virus (IAV) is a persistent threat to global health, causing seasonal epidemics and occasional pandemics that result in significant morbidity and mortality. This negative-sense single-stranded RNA virus belongs to the Orthomyxoviridae family and has a segmented genome encoding various essential proteins. In recent years, research has delved into the role of RNA modifications, particularly N6-methyladenosine (m6A), in the context of IAV infection. This blog post will explore the intricate relationship between m6A methylation and IAV, highlighting the latest scientific insights.
The Impact of Influenza A Virus
IAV primarily targets the respiratory system, and its consequences can range from mild flu symptoms to severe and life-threatening conditions. Severe cases may progress to acute respiratory distress syndrome (ARDS), septic shock, and multiple organ failure. The World Health Organization (WHO) estimates that influenza causes approximately 1 billion infections annually, with 3-5 million severe cases and up to 650,000 deaths from respiratory illness. The virus’s ability to mutate rapidly poses challenges for vaccine development, as current vaccines may not effectively protect against newly emerging strains. Additionally, the development of drug resistance among IAV strains further complicates the management of influenza infections.
Understanding N6-methyladenosine (m6A) Modification
m6A is one of the most prevalent and abundant post-transcriptional modifications in eukaryotes. It involves the addition of a methyl group to the N6 position of adenosine in RNA. This modification is dynamic and reversible, regulated by three key types of proteins:
- Methyltransferases (Writers): Responsible for installing the m6A mark. The core components of the m6A methyltransferase complex are METTL3 and METTL14, which form a stable heterodimer. Other associated proteins, such as WTAP, HAKAI, RBM15, RBM15B, VIRMA, and ZC3H13, help guide the complex to its target RNAs.
- Demethylases (Erasers): FTO and ALKBH5 are the two main m6A demethylases. They can remove the methyl group from m6A, thus reversing the modification.
- m6A-binding Proteins (Readers): These proteins recognize and bind to m6A-modified RNAs, mediating various downstream functions. Examples include the YTH family proteins (YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2), IGF2BP1, IGF2BP2, IGF2BP3, FMRP, and PRRC2A.
m6A modification impacts multiple aspects of RNA biology, including splicing, nuclear export, stability, and translation. It has been detected not only in mRNAs but also in non-coding RNAs such as lncRNAs, circRNAs, miRNAs, and more.
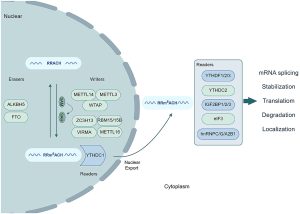
Fig.1 Mechanism of m6A modification.1
m6A Modification in the Context of IAV Infection
m6A in IAV Genomic RNAs
The initial discovery of m6A residues in IAV was in the hemagglutinin (HA) mRNA, which was found to have eight m6A modification sites. Since then, studies have shown that m6A modification is widespread among different influenza virus mRNAs, and the distribution of these modification sites varies. For example, recent research using m6A RNA immunoprecipitation sequencing (meRIP-seq) has revealed that multiple subtypes of IAV, including H5N6, H1N1, and H7N9, have m6A modifications on their viral RNA (vRNA), complementary RNA (cRNA), and mRNA. These modifications are conservatively distributed on the negative strands of genes such as NP, NA, and PB1.
Role of m6A in IAV Replication
Accumulating evidence suggests that m6A modification plays a significant role in regulating IAV replication. Knockdown of the methyltransferases METTL3/14 leads to a substantial decrease in the virus titer of H5N6, indicating that these enzymes are crucial for viral replication. Conversely, overexpression of METTL3/14 enhances the efficiency of viral RNA synthesis and increases the activity of the viral RNA-dependent RNA polymerase (RdRP). Specific mutations in the m6A motifs on the negative strands of NP, NA, and PB1 genes can reduce the m6A level on the mutant virus RNA, accompanied by a decrease in viral protein expression.
Impact on Viral Protein-RNA Interactions
m6A modification affects the interaction between viral proteins and RNAs. RNA-protein interaction experiments, such as cross-linking immunoprecipitation (CLIP) and RNA antisense purification (RAP), have demonstrated that the absence of m6A on NP vRNA reduces its binding efficiency to PB2/PB1/PA/NP proteins by about 70%, resulting in defective assembly of the viral ribonucleoprotein (vRNP) complex. This highlights the importance of m6A in maintaining the proper structure and function of the vRNP complex, which is essential for viral replication and transcription.
m6A and Host Immune Response
During IAV infection, the host immune system is activated in response to viral components. m6A modification can influence both the virus and the host immune response. Although m6A-deficient vRNA shows an increased binding to RIG-I (a key pattern recognition receptor in the host), the induction of IFN-β and ISRE (interferon-stimulated response element) activities by virus infection does not show significant differences. This suggests that the impact of m6A on the host immune response during IAV infection is complex and may involve multiple factors.
Therapeutic Implications
The discovery of the role of m6A in IAV infection opens up new avenues for developing antiviral strategies. Since m6A modification is crucial for IAV replication, targeting the m6A regulatory machinery could be a potential approach. For example, inhibiting the activity of methyltransferases like METTL3/14 or activating demethylases such as ALKBH5/FTO may disrupt the m6A-mediated viral replication process. Some small-molecule compounds, like STM2457, which has shown inhibitory effects on METTL3 in tumor models, could potentially be explored for their antiviral properties. Additionally, understanding how m6A affects the host immune response to IAV may also lead to the development of immunomodulatory therapies that enhance the host’s ability to clear the virus.
Latest Research Updates
Recent studies continue to uncover new details about the m6A – IAV relationship. For instance, research is focusing on identifying more precisely how different m6A – binding proteins interact with IAV RNAs and what impact this has on viral life cycle stages. There is also an increasing interest in understanding how m6A modification patterns change during the course of IAV infection in different host cell types. Furthermore, efforts are underway to develop more specific and effective inhibitors targeting the m6A regulatory proteins for potential antiviral drug development.
In conclusion, the study of m6A methylation in the context of IAV infection has provided valuable insights into the virus – host interaction. As we continue to explore this area, we can expect to develop more targeted and effective strategies to combat influenza, which remains a significant global health concern.
Creative Biolabs offers comprehensive mRNA modification services designed to enhance mRNA stability, reduce immunogenicity, and improve translational efficiency for therapeutic and research applications. Leveraging advanced technologies and expertise in in vitro transcription (IVT) and nucleic acid chemistry, the company provides customizable solutions tailored to client needs, including 5′ capping, nucleoside modification, and functional labeling.
5′ End Capping Modification Services
To protect mRNA from degradation, promote translation initiation, and regulate nuclear export by modifying the 5′ cap structure.
- mRNA 5′ Capping Services
Adds natural or synthetic cap structures (e.g., Cap 0, Cap 1, Cap 2) to mimic eukaryotic mRNA processing, enhancing stability and translation efficiency. - ARCAs Cap Service
Uses anti-reverse cap analogs (ARCAs) to ensure correct cap orientation during IVT, resulting in higher translation efficiency and resistance to decapping enzymes. - Fluorescent Cap Service
Incorporates fluorescent labels (e.g., anthraniloyl, trinitrophenyl) into the 5′ cap for real-time visualization and tracking of mRNA in cellular studies via fluorescence microscopy. - Fluorophosphate-containing Cap Service
Integrates fluorophosphate moieties into the cap to monitor enzyme activity (e.g., decapping enzymes) and study protein-RNA interactions, enhancing analytical capabilities. - 6-Thioguanosine-containing Cap Service
Introduces 6-thioguanosine (s6G) for photo-crosslinking studies, improving mRNA stability and enabling precise mapping of RNA-protein interactions.
Nucleotide Base Modification Services
To modify nucleoside bases (uridine, cytidine, adenosine) to enhance RNA structure, reduce immune response, and regulate gene expression.
- Nucleotides Modification Services
Provides custom modification of nucleosides (e.g., pseudouridine, 5-methylcytidine) to optimize mRNA stability, translation efficiency, and functional diversity for therapeutic applications. - Pseudouridine Modification Service
Substitutes uridine with pseudouridine (Ψ) to stabilize RNA secondary structures, improve translational accuracy, and minimize immune activation. - 2-Thiouridine Modification Service
Replaces oxygen with sulfur at the C-2 position of uridine to enhance tRNA stability, thermal resistance, and translation fidelity. - 5-Methylcytidine Modification Service
Adds a methyl group to cytidine (m⁵C) to regulate mRNA decay, splicing, and epigenetic processes, with applications in stem cell differentiation and disease research. - N6-Methyladenosine Modification Service
Introduces m⁶A, the most abundant mRNA modification, to regulate splicing, nuclear export, and translation, with advanced detection methods (e.g., MeRIP-seq, HPLC).
References
- Liu, Xueer, et al. “RNA N6-methyladenosine methylation in influenza A virus infection.” Frontiers in Microbiology15 (2024): 1401997. https://doi.org/10.3389/fmicb.2024.1401997
